

Sequencher contig windows#
Rearrange the windows so that you optimize the view of the Chromatograms, the Contig Editor, and the Variance Table. You can also customize the layout of the windows, so that Sequencher will remember how you like to work.
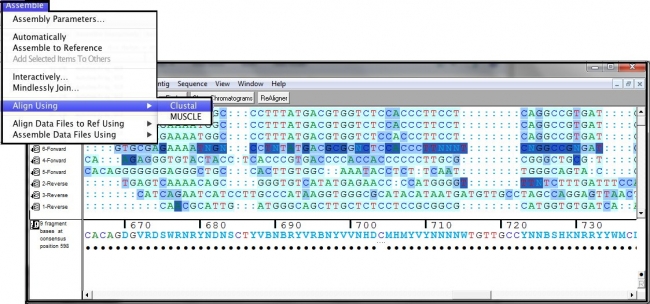
This opens the Variance Table in Review mode so that you can now see all of the data that supports each difference reported in the table.
Sequencher contig full#
However, the fact that each of the cells that contain the name of a contig is shaded in pink indicates that the data in the contigs does not cover the full length of the rCRS. Every difference that is reported in one is confirmed in the other. The sample sequences differ from the Reference Sequence at positions 73 and 195. Each column represents a sample sequence. In the bottom left corner of the Variance Table, click on the resize icon button to optimize the column widths for the display of the full names of the contigs.Įach row in the Variance Table represents a position in the Reference Sequence that has a difference in one of the sample sequences. Select both contigs and then select Contig > Compare Consensus to Reference. The process of editing and assembling sample data into a contig is described in detail in the Reference Sequence Tutorial.ĬOMPARING THE DATA Because both contigs are assembled to the same Reference Sequence, the rCRS, it is possible to compare equivalent base positions. The two imported contigs contain data from the same sample, but it was sequenced in two different labs. Repeat to import 082790.SPF and click Open. Choose the project 90.SPF and click Open. mtDNA Profile projects are located in the Mitochondrial Sequences/mtDNA Profiles folder. įrom the File menu, choose Import > Sequencher Project… Navigate to the Sample Data folder inside the Sequencher application folder. IMPORT SAMPLE DATA Now that you have your reference sequence you are ready to import your sample data.
Sequencher contig code#
In most instances the entire mitochondrial genome is not used for comparison, but just the region around the Dloop (16024-576), the control region that does not code for proteins. The accepted standard sequence for comparison is the revised Cambridge Reference Sequence, rCRS. An mtDNA profile is a list of the differences between the mtDNA sequence of a forensic sample and the sequence of a standard reference. It is the standard for mtDNA analysis in forensic laboratories across the country. The forensic version of Sequencher has specific functionality to meet the needs of forensic scientists analyzing mitochondrial (mtDNA) sequence.
Mitochondrial DNA Typing Tutorial There are two versions of Sequencher, the standard version, which has become the favorite for sequence analysis since its release in 1991, and the forensic version. 4 Comparing the Data.4 Validate mtDNA Profiles.7 Create an Analyst Report. Mitochondrial DNA Typing Open a Project Template.


 0 kommentar(er)
0 kommentar(er)
